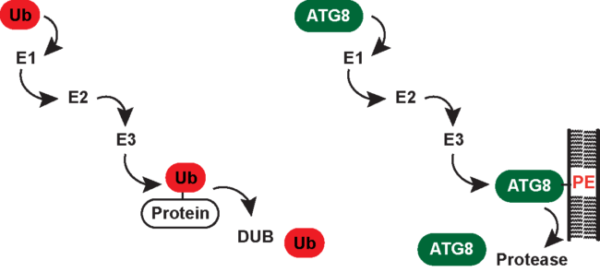
Covalent attachment of ubiquitin (Ub) to proteins serves as a versatile regulatory signal and controls the stability, localization or activation status of myriad cellular proteins. Ubiquitination occurs through isopeptide bond formation between the ε-amino group of a lysine residue in a target protein and the C-terminal carboxyl group of Ub. Proteins can be modified by Ub monomers or polymers. In the latter, Ub moieties are connected through lysine (Lys6, Lys11, Lys29, Lys33, Lys48 and Lys63)- and/or methionine (Met1)-mediated isopeptide linkages. Distinct classes of poly- and monoubiquitin signals are decoded by a variety of Ub-binding proteins that help exerting their biological functions such as proteasomal degradation in the case of Lys48 linkage. Ub chain synthesis involves an enzymatic E1-E2-E3 cascade in which the E3 Ub ligase recognizes the substrate and brings it into proximity of the E2 Ub-conjugating enzyme. Deubiquitinating enzymes (DUBs) function to disassemble Ub chains and can either act to reverse ubiquitination of proteins destined for degradation by the proteasome or remove regulatory Ub modifications in order to turn a particular pathway on or off. In contrast, the Ub-like protein ATG8 is uniquely conjugated to the phospholipid phosphatidylethanolamine (PE). Reversible PE-modification is achieved through concerted action of ATG8-specific E1, E2, E3 and DUB enzymes. Human cells contain six ATG8 family members, namely, microtubule-associated protein 1A/1B light chain 3A (LC3A), LC3B, LC3C, γ-aminobutyric acid receptor-associated protein (GABARAP), GABARAPL1 and GABARAPL2. A variety of proteins bind human ATG8 proteins (hATG8s) via a short linear sequence known as the LC3-interaction region (LIR). The best-understood function of hATG8s is in autophagy, which is a membrane-based degradation process that enables cells to engulf a portion of their cytoplasm and deliver it to lysosomes for bulk degradation. Here, hATG8-PE conjugates are incorporated into both layers of incipient autophagosomes where hATG8s provide a docking site for autophagy cargo receptors (e.g. p62 and OPTN) and regulatory proteins, which control formation, maturation and transport of autophagosomes as well as their fusion with lysosomes.
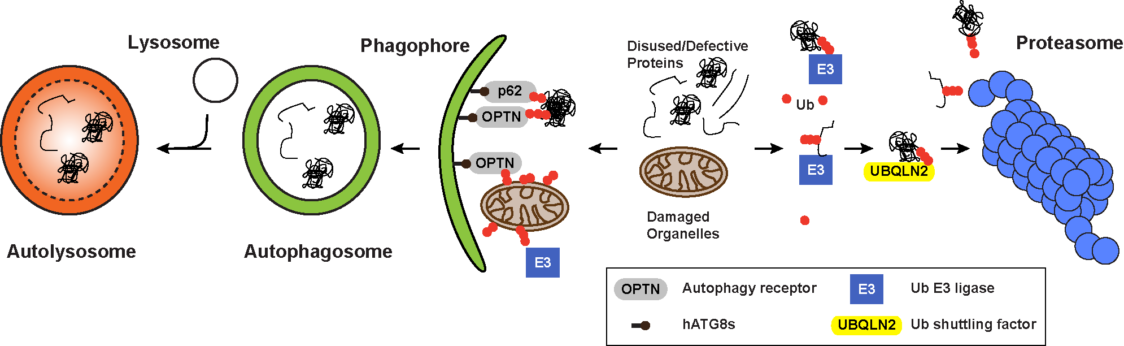
Both the autophagy and ubiquitin systems have evolved to maintain a functional proteome which is a key requirement for cell metabolism, organelle biogenesis, stress adaptation and consequently the long-term viability of any cell type and organ. A major challenge in proteostasis concerns protection against the detrimental consequences of unfolded, misfolded or damaged proteins that severely disturb cellular functions and are associated with aging and age-related diseases, including neurodegeneration, most of which are currently incurable. While the Ub-proteasome system (UPS) is the primary proteolytic route for short-lived, misfolded and damaged proteins, autophagy is the preferred degradative route for large, heterogeneous cytoplasmic materials whose sizes exceed the spatial capacity of proteasomes. Under nutrient or growth factor deprivation, autophagy is thought to be nonselective and dedicated to bulk degradation of any cytosolic protein and other macromolecules to deliver essential nutrients. However, autophagy can also selectively target cellular structures including damaged organelles such as mitochondria (mitophagy), protein aggregates, stress granules and invading bacteria (xenophagy). In this context, selective autophagy functions as an important cytoprotective mechanism that can be activated in nutrient-rich conditions by various stress signals. Both pathways, autophagy and the UPS, are interconnected and communicate with each other. However, one of the most striking commonality of both degradation systems is the use of ubiquitylation as a labeling system for their substrates.
Work in our lab centers on advancing our understanding of how autophagy and ubiquitin contribute to maintain proteostasis. In particular, we are interested in defining components, targets, regulators and crosstalk of both pathways. The lab is driven by the principle of coupling systematic, unbiased approaches alongside with functional and mechanistic analyses to test hypotheses across a multiple of biological systems and models. This approach allowed us to make several important discoveries with regard to novel
i) Autophagy and ubiquitin pathway components
By employing an image-based screening approach monitoring multiple autophagosome markers at endogenous levels we identified novel autophagy regulators among the families of Rab GTPases and their GEF and GAP regulators. For SMCR8 we revealed an unconventional nuclear function in controlling autophagic gene expression (Jung et al. 2017). Additionally, we identified a so far unknown molecular link between endosomes and autophagosomes by functionally characterizing the LC3-binding autophagy adapter TBC1D5 (Popovic et al. 2012). In our systematic proteomic analysis of the human autophagy system, we identified a large cohort of proteins that associate with LC3 and GABARAP proteins, with extensive binding partner overlap between family members, and frequent involvement of the conserved surface on hATG8 proteins known to interact with LIR motifs in partner proteins. This plethora of proteins is likely to contain within it the seeds to understanding the diversity of the human ATG8 family (Behrends et al. 2010). Moreover, we identified ARIH1 as one of the E3 ligases involved in the formation of ubiquitin coat on cytosolic S. Typhimurium. Together with LRSAM1 and HOIP, ARIH1 forms a network of E3 ligases that recognize cytosolic bacteria and mediate xenophagic degradation and host immune response (Polajnar et al. 2017).
ii) Autophagy cargo and ubiquitylation substrates
We employed proximity proteomics-based autophagosome content profiling to identified a role for the ATG8 family member LC3C in maintaining basal mitochondrial homeostasis. Selected mitochondrial proteins among them MTX1 were targeted by LC3C and p62 through a piece-meal mitophagy pathway (Le Guerroué et al. 2017). By quantitatively profiling the ubiquitinome of Salmonella-infected epithelial cells, our comprehensive analysis highlighted the ubiquitination-mediated regulation of CDC42 and NFκB activities, revealed novel host targets of bacterial E3 ligases/DUBs and demonstrated extensive ubiquitination of bacterial effectors and outer membrane proteins (Fishkin et al. 2016).
iii) Regulators of autophagy and ubiquitin pathways
We revealed a function for TECPR2 (a hereditary spastic paraplegias-linked ATG8-interacting protein) that binds to the ATG8 family member LC3C and maintains endoplasmic reticulum export, thereby critically contributing to autophagosome formation (Stadel et al. 2015). By eliciting upstream signaling cascades regulating autophagy, we identified SLC38A9 as a novel component of the Rag-Ragulator complex that helps transducing amino acid availability to mTORC1 activity (Jung et al. 2015). In a collaborative effort, we identified a function of the enigmatic pseudophosphatase STYX as novel regulator of cullin RING ubiquitin ligases (Reiterer et al., 2017).
iv) Crosstalk between autophagy and ubiquitin pathways
We identified novel functions for the ATG8 subfamily of GABARAP proteins going beyond autophagy in providing spatio-temporal control of RAC1 signaling through a cullin RING ubiquitin E3 ligase (Genau et al. 2015).